英文名稱PCNA (Nuclear Loading Control)
中文名稱增殖細(xì)胞核抗原(核內(nèi)參)重組兔單克隆抗體
別 名Cyclin; DNA polymerase delta auxiliary protein; HGCN8729; MGC8367; Mutagen-sensitive 209 protein; Pcna]9PGF.gif) clin; PCNAR; Polymerase delta accessory protein; Proliferating Cell Nuclear Antigen; PCNA_HUMAN.
clin; PCNAR; Polymerase delta accessory protein; Proliferating Cell Nuclear Antigen; PCNA_HUMAN.
產(chǎn)品類型內(nèi)參抗體
研究領(lǐng)域腫瘤 細(xì)胞生物 染色質(zhì)和核信號(hào) 細(xì)胞周期蛋白
抗體來源Rabbit
克隆類型Monoclonal
克 隆 號(hào)1G3
交叉反應(yīng)Human, Mouse, Rat,
產(chǎn)品應(yīng)用WB=1:1000-5000 IP=1:10-50 IHC-P=1:100-500 IHC-F=1:100-500 ICC=1:50-200 IF=1:100-500 (石蠟切片需做抗原修復(fù))
not yet tested in other applications.
optimal dilutions/concentrations should be determined by the end user.
分 子 量29kDa
細(xì)胞定位細(xì)胞核
性 狀Liquid
濃 度1mg/ml
免 疫 原Recombinant human PCNA protein:
亞 型IgG
純化方法affinity purified by Protein A
儲(chǔ) 存 液0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol.
保存條件Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles.
PubMedPubMed
產(chǎn)品介紹Proliferating cell nuclear antigen (PCNA) is a 28kDa nuclear protein associated with the cell cycle, a nuclear protein vital for cellular DNA synthesis. Proliferating cell nuclear antigen was originally identified by immunofluorescence as a nuclear protein whose appearance correlated with the proliferate state of the cell. PCNA is required for replication of DNA in vitro and has been identified as the auxiliary protein (cofactor) for DNA polymerase delta. The anti-PCNA antibodies react with the nuclei of proliferating cells. PCNA is essential for cellular DNA synthesis and is also required for the in vitro replication of simian virus 40 (SV40) DNA where it acts to coordinate leading and lagging strand synthesis at the replication fork. The PCNA protein may fulfil several separate roles in the cell nucleus associated with changes in its antigenic structure.
Function:
Auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand. Induces a robust stimulatory effect on the 3'-5' exonuclease and 3'-phosphodiesterase, but not apurinic-apyrimidinic (AP) endonuclease, APEX2 activities. Has to be loaded onto DNA in order to be able to stimulate APEX2. Plays a key role in DNA damage response (DDR) by being conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways. Acts as a loading platform to recruit DDR proteins that allow completion of DNA replication after DNA damage and promote postreplication repair: Monoubiquitinated PCNA leads to recruitment of translesion (TLS) polymerases, while 'Lys-63'-linked polyubiquitination of PCNA is involved in error-free pathway and employs recombination mechanisms to synthesize across the lesion.
Subunit:
Homotrimer. Forms a complex with activator 1 heteropentamer in the presence of ATP. Interacts with EXO1, POLH, POLK, DNMT1, ERCC5, FEN1, CDC6 and POLDIP2. Interacts with APEX2; this interaction is triggered by reactive oxygen species and increased by misincorporation of uracil in nuclear DNA. Forms a ternary complex with DNTTIP2 and core histone. Interacts with KCTD10 and PPP1R15A (By similarity). Interacts with POLD1, POLD3 and POLD4. Interacts with BAZ1B; the interaction is direct. Interacts with HLTF and SHPRH. Interacts with NUDT15. Interaction is disrupted in response to UV irradiation and acetylation. Interacts with CDKN1A/p21(CIP1) and CDT1; interacts via their PIP-box which also recruits the DCX(DTL) complex. Interacts with DDX11. Interacts with EGFR; positively regulates PCNA. Interacts with PARPBP. Interacts (when ubiquitinated) with SPRTN; leading to enhance RAD18-mediated PCNA ubiquitination. Interacts (when polyubiquitinated) with ZRANB3. Interacts with SMARCAD1. Interacts with CDKN1C. Interacts with KIAA0101/PAF15 (via PIP-box).
Subcellular Location:
Nucleus. Note=Forms nuclear foci representing sites of ongoing DNA replication and vary in morphology and number during S phase. Together with APEX2, is redistributed in discrete nuclear foci in presence of oxidative DNA damaging agents.
Post-translational modifications:
Following DNA damage, can be either monoubiquitinated to stimulate direct bypass of DNA lesions by specialized DNA polymerases or polyubiquitinated to promote recombination-dependent DNA synthesis across DNA lesions by template switching mechanisms. Following induction of replication stress, monoubiquitinated by the UBE2B-RAD18 complex on Lys-164, leading to recruit translesion (TLS) polymerases, which are able to synthesize across DNA lesions in a potentially error-prone manner. An error-free pathway also exists and requires non-canonical polyubiquitination on Lys-164 through 'Lys-63' linkage of ubiquitin moieties by the E2 complex UBE2N-UBE2V2 and the E3 ligases, HLTF, RNF8 and SHPRH. This error-free pathway, also known as template switching, employs recombination mechanisms to synthesize across the lesion, using as a template the undamaged, newly synthesized strand of the sister chromatid. Monoubiquitination at Lys-164 also takes place in undamaged proliferating cells, and is mediated by the DCX(DTL) complex, leading to enhance PCNA-dependent translesion DNA synthesis. Sumoylated during S phase.
Acetylated in response to UV irradiation. Acetylation disrupts interaction with NUDT15 and promotes degradation.
Phosphorylated. Phosphorylation at Tyr-211 by EGFR stabilizes chromatin-associated PCNA.
Similarity:
Belongs to the PCNA family.
SWISS:
P12004
Gene ID:
5111
Database links:
Entrez Gene: 515499 Cow
Entrez Gene: 5111 Human
Entrez Gene: 18538 Mouse
Entrez Gene: 25737 Rat
Omim: 176740 Human
SwissProt: Q3ZBW4 Cow
SwissProt: P12004 Human
SwissProt: P17918 Mouse
SwissProt: P04961 Rat
Unigene: 147433 Human
Unigene: 728886 Human
Unigene: 7141 Mouse
Unigene: 223 Rat
Important Note:
This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications.
別 名Cyclin; DNA polymerase delta auxiliary protein; HGCN8729; MGC8367; Mutagen-sensitive 209 protein; Pcna
]9PGF.gif) clin; PCNAR; Polymerase delta accessory protein; Proliferating Cell Nuclear Antigen; PCNA_HUMAN.
clin; PCNAR; Polymerase delta accessory protein; Proliferating Cell Nuclear Antigen; PCNA_HUMAN. 產(chǎn)品類型內(nèi)參抗體
研究領(lǐng)域腫瘤 細(xì)胞生物 染色質(zhì)和核信號(hào) 細(xì)胞周期蛋白
抗體來源Rabbit
克隆類型Monoclonal
克 隆 號(hào)1G3
交叉反應(yīng)Human, Mouse, Rat,
產(chǎn)品應(yīng)用WB=1:1000-5000 IP=1:10-50 IHC-P=1:100-500 IHC-F=1:100-500 ICC=1:50-200 IF=1:100-500 (石蠟切片需做抗原修復(fù))
not yet tested in other applications.
optimal dilutions/concentrations should be determined by the end user.
分 子 量29kDa
細(xì)胞定位細(xì)胞核
性 狀Liquid
濃 度1mg/ml
免 疫 原Recombinant human PCNA protein:
亞 型IgG
純化方法affinity purified by Protein A
儲(chǔ) 存 液0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol.
保存條件Shipped at 4℃. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles.
PubMedPubMed
產(chǎn)品介紹Proliferating cell nuclear antigen (PCNA) is a 28kDa nuclear protein associated with the cell cycle, a nuclear protein vital for cellular DNA synthesis. Proliferating cell nuclear antigen was originally identified by immunofluorescence as a nuclear protein whose appearance correlated with the proliferate state of the cell. PCNA is required for replication of DNA in vitro and has been identified as the auxiliary protein (cofactor) for DNA polymerase delta. The anti-PCNA antibodies react with the nuclei of proliferating cells. PCNA is essential for cellular DNA synthesis and is also required for the in vitro replication of simian virus 40 (SV40) DNA where it acts to coordinate leading and lagging strand synthesis at the replication fork. The PCNA protein may fulfil several separate roles in the cell nucleus associated with changes in its antigenic structure.
Function:
Auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand. Induces a robust stimulatory effect on the 3'-5' exonuclease and 3'-phosphodiesterase, but not apurinic-apyrimidinic (AP) endonuclease, APEX2 activities. Has to be loaded onto DNA in order to be able to stimulate APEX2. Plays a key role in DNA damage response (DDR) by being conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways. Acts as a loading platform to recruit DDR proteins that allow completion of DNA replication after DNA damage and promote postreplication repair: Monoubiquitinated PCNA leads to recruitment of translesion (TLS) polymerases, while 'Lys-63'-linked polyubiquitination of PCNA is involved in error-free pathway and employs recombination mechanisms to synthesize across the lesion.
Subunit:
Homotrimer. Forms a complex with activator 1 heteropentamer in the presence of ATP. Interacts with EXO1, POLH, POLK, DNMT1, ERCC5, FEN1, CDC6 and POLDIP2. Interacts with APEX2; this interaction is triggered by reactive oxygen species and increased by misincorporation of uracil in nuclear DNA. Forms a ternary complex with DNTTIP2 and core histone. Interacts with KCTD10 and PPP1R15A (By similarity). Interacts with POLD1, POLD3 and POLD4. Interacts with BAZ1B; the interaction is direct. Interacts with HLTF and SHPRH. Interacts with NUDT15. Interaction is disrupted in response to UV irradiation and acetylation. Interacts with CDKN1A/p21(CIP1) and CDT1; interacts via their PIP-box which also recruits the DCX(DTL) complex. Interacts with DDX11. Interacts with EGFR; positively regulates PCNA. Interacts with PARPBP. Interacts (when ubiquitinated) with SPRTN; leading to enhance RAD18-mediated PCNA ubiquitination. Interacts (when polyubiquitinated) with ZRANB3. Interacts with SMARCAD1. Interacts with CDKN1C. Interacts with KIAA0101/PAF15 (via PIP-box).
Subcellular Location:
Nucleus. Note=Forms nuclear foci representing sites of ongoing DNA replication and vary in morphology and number during S phase. Together with APEX2, is redistributed in discrete nuclear foci in presence of oxidative DNA damaging agents.
Post-translational modifications:
Following DNA damage, can be either monoubiquitinated to stimulate direct bypass of DNA lesions by specialized DNA polymerases or polyubiquitinated to promote recombination-dependent DNA synthesis across DNA lesions by template switching mechanisms. Following induction of replication stress, monoubiquitinated by the UBE2B-RAD18 complex on Lys-164, leading to recruit translesion (TLS) polymerases, which are able to synthesize across DNA lesions in a potentially error-prone manner. An error-free pathway also exists and requires non-canonical polyubiquitination on Lys-164 through 'Lys-63' linkage of ubiquitin moieties by the E2 complex UBE2N-UBE2V2 and the E3 ligases, HLTF, RNF8 and SHPRH. This error-free pathway, also known as template switching, employs recombination mechanisms to synthesize across the lesion, using as a template the undamaged, newly synthesized strand of the sister chromatid. Monoubiquitination at Lys-164 also takes place in undamaged proliferating cells, and is mediated by the DCX(DTL) complex, leading to enhance PCNA-dependent translesion DNA synthesis. Sumoylated during S phase.
Acetylated in response to UV irradiation. Acetylation disrupts interaction with NUDT15 and promotes degradation.
Phosphorylated. Phosphorylation at Tyr-211 by EGFR stabilizes chromatin-associated PCNA.
Similarity:
Belongs to the PCNA family.
SWISS:
P12004
Gene ID:
5111
Database links:
Entrez Gene: 515499 Cow
Entrez Gene: 5111 Human
Entrez Gene: 18538 Mouse
Entrez Gene: 25737 Rat
Omim: 176740 Human
SwissProt: Q3ZBW4 Cow
SwissProt: P12004 Human
SwissProt: P17918 Mouse
SwissProt: P04961 Rat
Unigene: 147433 Human
Unigene: 728886 Human
Unigene: 7141 Mouse
Unigene: 223 Rat
Important Note:
This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications.
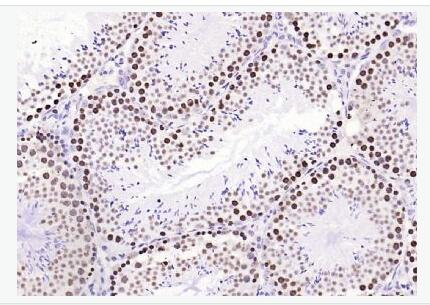
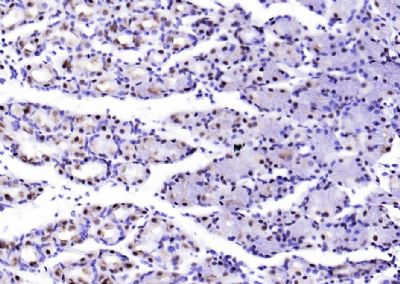
| 產(chǎn)品圖片 | 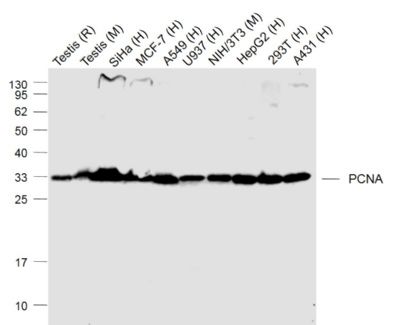 Sample: Lane 1: Testis (Rat) Lysate at 40 ug Lane 2: Testis (Mouse) Lysate at 40 ug Lane 3: SiHa (Human) Cell Lysate at 30 ug Lane 4: MCF-7 (Human) Cell Lysate at 30 ug Lane 5: A549 (Human) Cell Lysate at 30 ug Lane 6: U937 (Human) Cell Lysate at 30 ug Lane 7: NIH/3T3(Mouse) Cell Lysate at 30 ug Lane 8: HepG2 (Human) Cell Lysate at 30 ug Lane 9: 293T (Human) Cell Lysate at 30 ug Lane 10: A431 (Human) Cell Lysate at 30 ug Primary: Anti-PCNA (bsm-52347R) at 1/1000 dilution Secondary: IRDye800CW Goat Anti-Rabbit IgG at 1/20000 dilution Predicted band size: 32’36 kD Observed band size: 32 kD 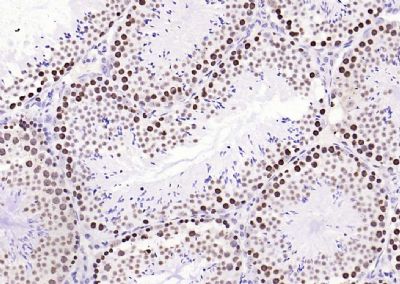 Paraformaldehyde-fixed, paraffin embedded (mouse testis); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 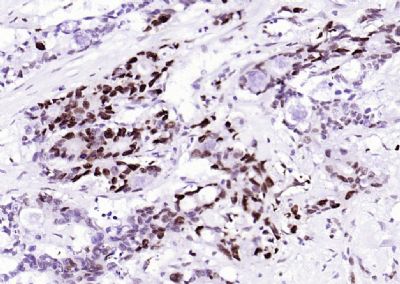 Paraformaldehyde-fixed, paraffin embedded (human rectal carcinoma); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 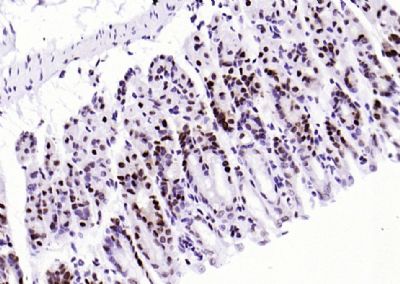 Paraformaldehyde-fixed, paraffin embedded (mouse stomach); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 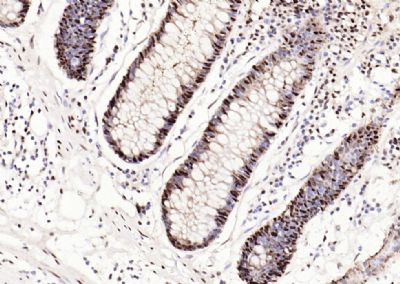 Paraformaldehyde-fixed, paraffin embedded (human rectal carcinoma); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 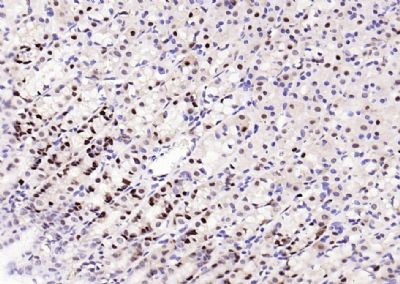 Paraformaldehyde-fixed, paraffin embedded (rat stomach); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 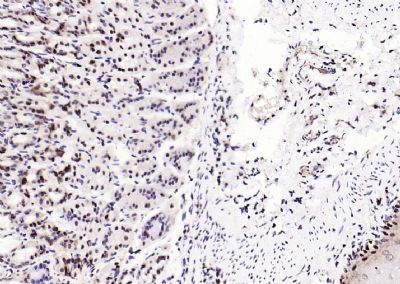 Paraformaldehyde-fixed, paraffin embedded (mouse stomach); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 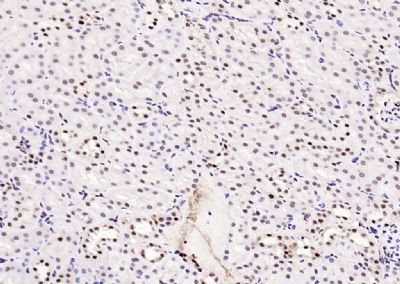 Paraformaldehyde-fixed, paraffin embedded (rat kidney); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. Paraformaldehyde-fixed, paraffin embedded (human gastric carcinoma); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 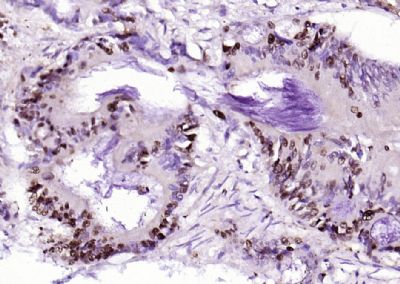 Paraformaldehyde-fixed, paraffin embedded (human cervical carcinoma); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 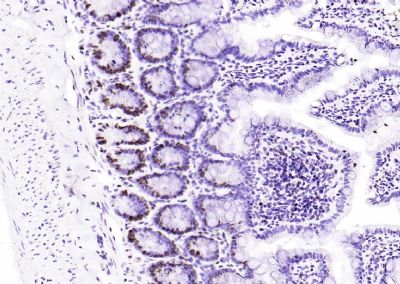 Paraformaldehyde-fixed, paraffin embedded (Mouse small intestine); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 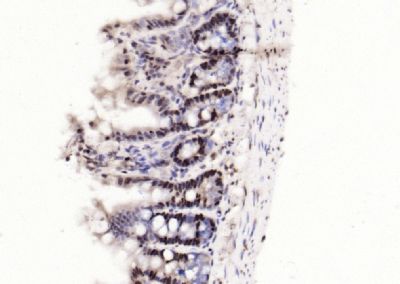 Paraformaldehyde-fixed, paraffin embedded (mouse small intestine); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 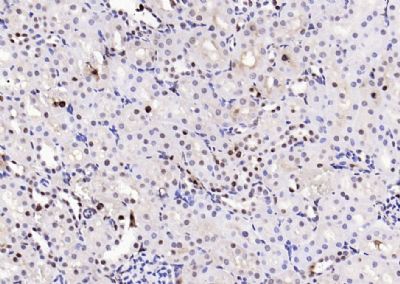 Paraformaldehyde-fixed, paraffin embedded (mouse kidney); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. 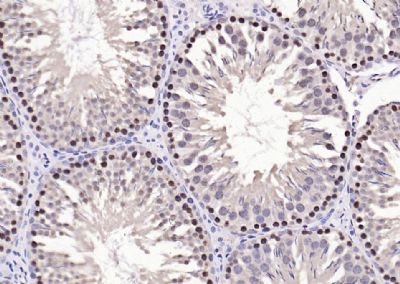 Paraformaldehyde-fixed, paraffin embedded (rat testis); Antigen retrieval by boiling in sodium citrate buffer (pH6.0) for 15min; Block endogenous peroxidase by 3% hydrogen peroxide for 20 minutes; Blocking buffer (normal goat serum) at 37°C for 30min; Antibody incubation with (PCNA (Nuclear Loading Control)) Monoclonal Antibody, Unconjugated (bsm-52347R) at 1:200 overnight at 4°C, followed by operating according to SP Kit(Rabbit) (sp-0023) instructionsand DAB staining. |
我要詢價(jià)
*聯(lián)系方式:
(可以是QQ、MSN、電子郵箱、電話等,您的聯(lián)系方式不會(huì)被公開)
*內(nèi)容: